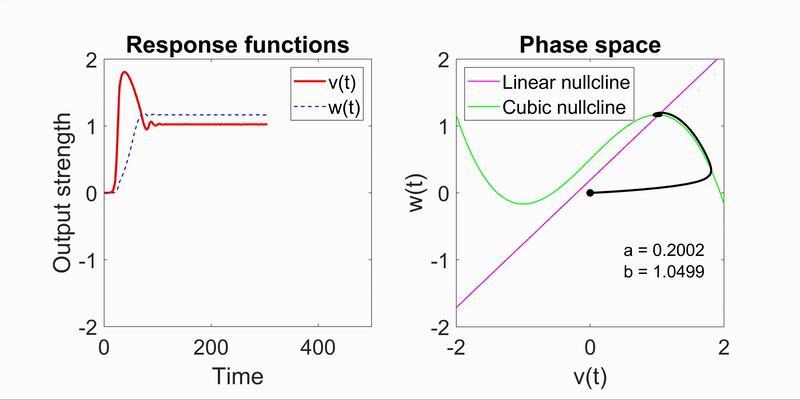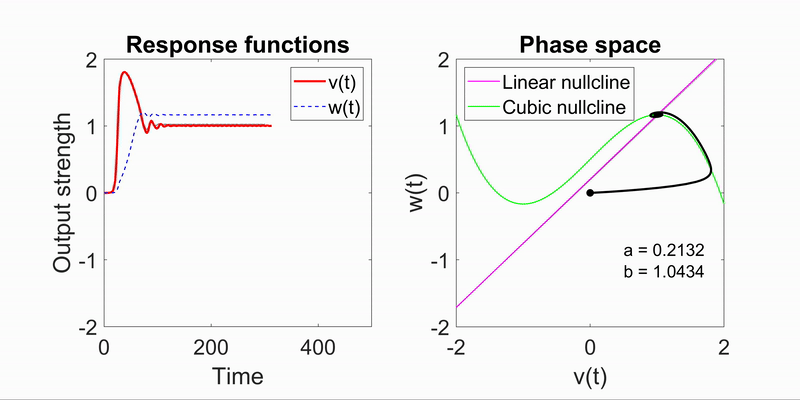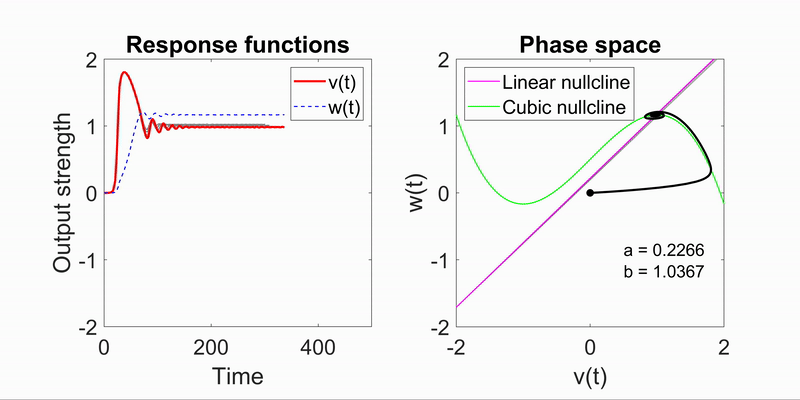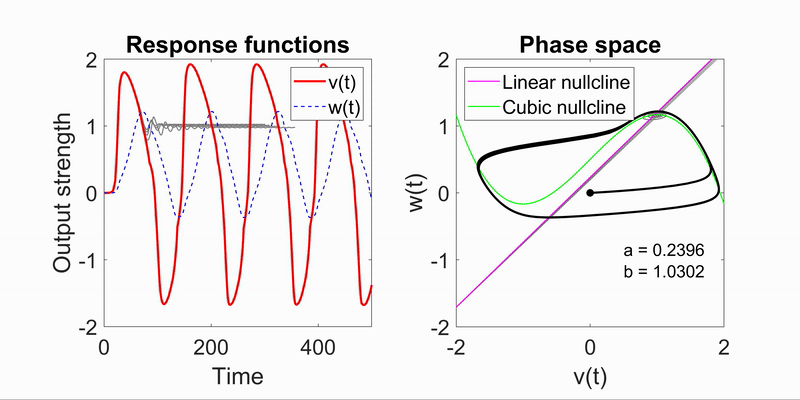
Modeling Flow-Field Dynamics in Biological Systems
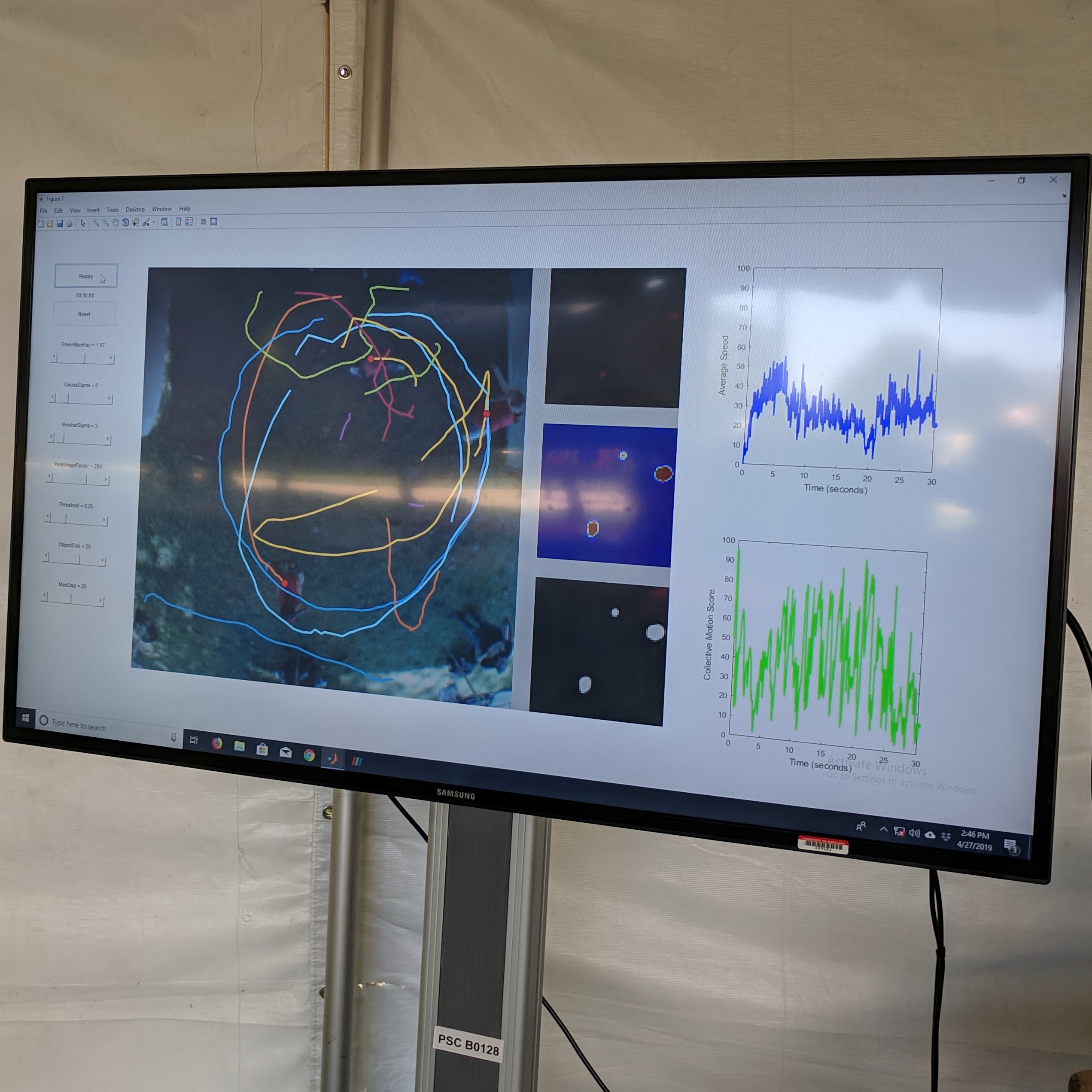
Optical flow measurements enable a wide variety of technologies, from facial recognition to self-driving cars. In this project, I designed an optical-flow-based pipeline to quantify flow-field dynamics in biological systems.
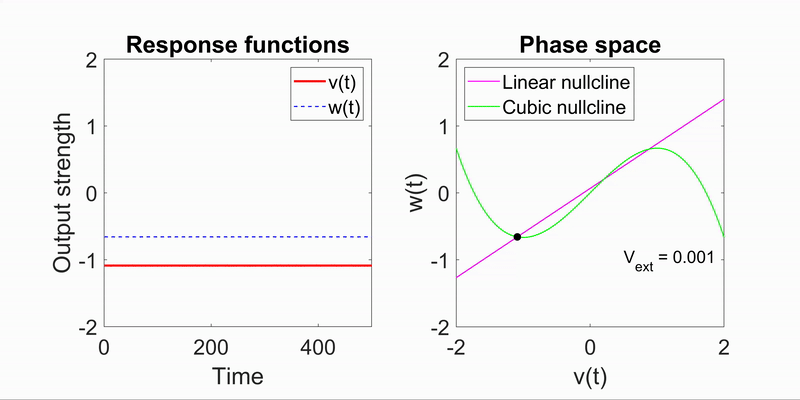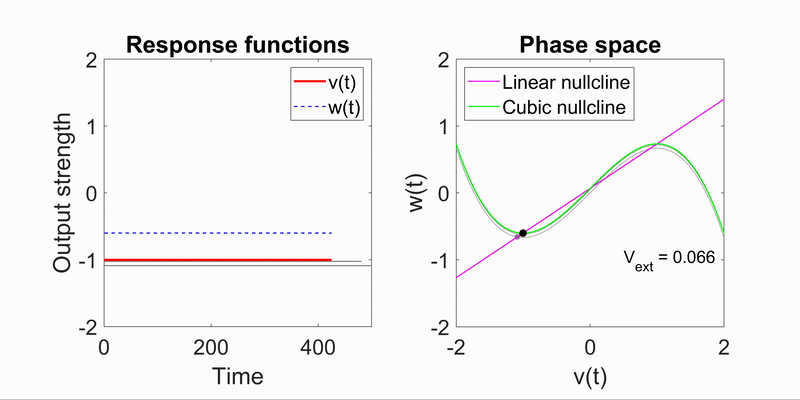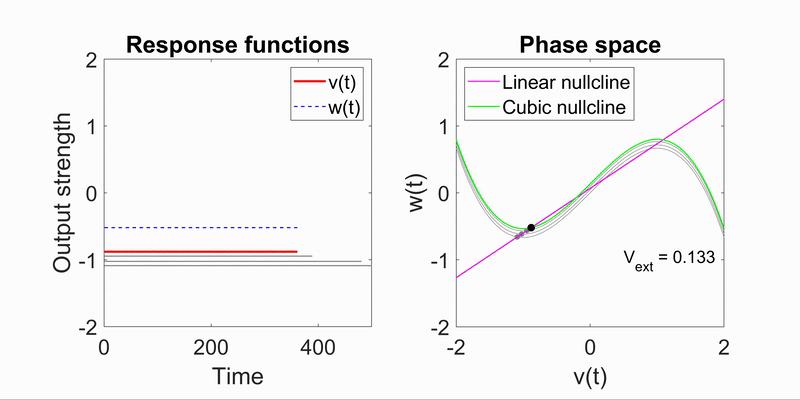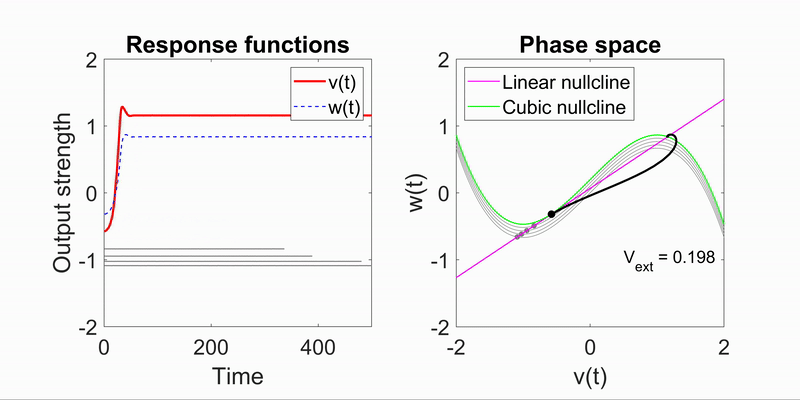
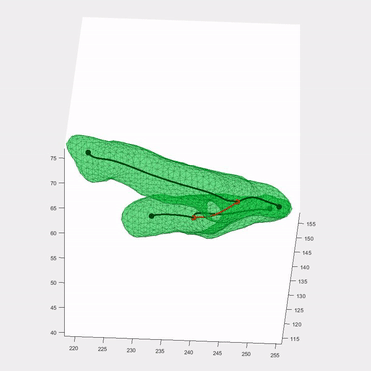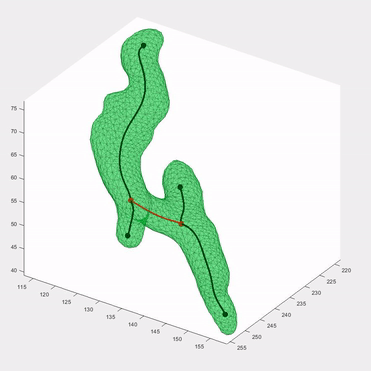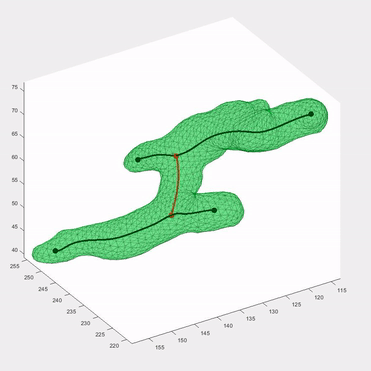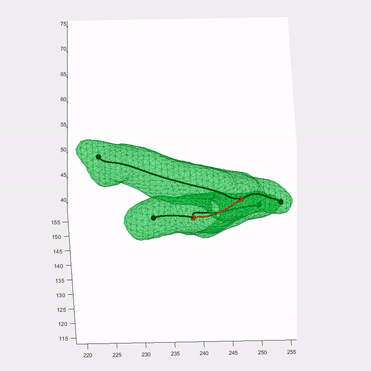
Applying the algorithm to microscope images, I extracted measurements such as speed and directionality across various cell types. I then modeled these flow fields using a bimodal mixed von-Mises distribution, capturing the influence of external forces on cellular behavior.
This algorithm was introduced in Lee*, Campanello*, et al., MBoC 2020 (preprint and manuscript), and was featured in several follow-up publications.
The original MATLAB code for the MBoC manuscript can be found at github.com/LosertLab/FlowClusterTracking, with updates at github.com/ljcamp1624/FlowClusterTracking.
Key concepts: computer vision, optimization, modeling, maximum-likelihood estimation, data visualization, time-series analysis.